Testing universal common ancestry?
Theobald has an interesting paper in this week's Nature, "A formal test of the theory of universal common ancestry." In it, he claims to have tested the idea that all organisms - bacteria, archaea, plants, animals, fungi, etc. - descended from a common ancestor. According to Theobald,
Among a wide range of biological models involving the independent ancestry of major taxonomic groups, the model selection tests are found to overwhelmingly support UCA irrespective of the presence of horizontal gene transfer and symbiotic fusion events. These results provide powerful statistical evidence corroborating the monophyly of all known life.Although Theobald does not cite creationists in the article, I think it's pretty clear who his primary target is.
There is much to be admired about the article. The fact that he admits that universal common ancestry has not been formally tested is a step in the right direction. He correctly lists most of the qualitative evidence in favor of universal common ancestry (with the exception of biogeography, which at most supports common ancestry at the level of order or maybe class). He also correctly recognizes that statistically significant sequence similarity is just an observation, while common ancestry (evolutionary "homology") is an interpretation. But he makes some strange claims about sequence similarity that I think warrant closer examination, especially since his results seem to be dependent on that similarity.
The first strange claim:
Statistically significant sequence similarity can arise from factors other than common ancestry, such as convergent evolution due to selection, structural constraints on sequence identity, mutation bias, chance, or artefact manufacture.On the surface, this is extremely unlikely, but it's hard to evaluate carefully since he doesn't define "statistically significant." Since I happen to know a little about sequence similarity, I can confidently affirm that the statistical significance estimates for BLAST or FASTA are extremely accurate, and if the expectation values are interpreted properly, the chance of getting a false positive is very low.
Where does Theobald get this idea? To support the claim, he cites a 1998 paper by Murzin, "How far divergent evolution goes in proteins," where Murzin cites convergence and structural constraints as sources of structural - not sequence - similarity. Indeed, Murzin affirms, "Strong sequence similarity alone is considered to be a sufficient evidence for the common ancestry," and the examples he discusses in the paper are proteins that have structural but not sequence similarity.
OK, so there's one dubious sentence in the introduction of the paper. So what? Well, Theobald also wants to claim that his formal test does not depend on sequence similarity indicating homology. In his words:
Here I report tests of the theory of UCA using model selection theory, without assuming that sequence similarity indicates a genealogical relationship.How does that work? For his data, he takes 23 "universally conserved" proteins that are found in twelve organisms from all three domains of life (archaea, bacteria, and eukaryotes). How did he know they were universally conserved? "significant sequence similarity using BLAST searches." That's an important point that I'll get back to below.
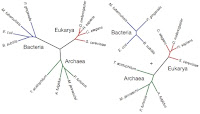
What I think he means by "without assuming ... a genealogical relationship" is that he explicitly modeled independent trees (see above). In the case on the left, he's modeling a common ancestry for all organisms in the dataset, while on the right he's put the bacteria on a separate tree from the eukaryotes and archaea. That's his model of independent origins, and he finds that "overwhelmingly" the selection criteria support models with a single tree rather than the models with multiple trees.
Why is that? According to Theobald,
What property of the sequence data supports common ancestry so decisively? When two related taxa are separated into two trees, the strong correlations that exist between the sequences are no longer modelled, which results in a large decrease in the likelihood. Consequently, when comparing a common-ancestry model to a multiple-ancestry model, the large test scores are a direct measure of the increase in our ability to accurately predict the sequence of a genealogically related protein relative to an unrelated protein."Strong correlations that exist between the sequences" = sequence similarity. How can we know this? Theobald continues,
The sequence correlations between a given clade of taxa and the rest of the tree would be eliminated if the columns in the sequence alignment for that clade were randomly shuffled. In such a case, these model-based selection tests should prefer the multiple-ancestry model. ...the multiple-ancestry models for shuffled data sets are preferred by a large margin over common ancestry models.Shuffling sequences eliminates sequence similarity, and therefore multiple-ancestry models are more likely than common ancestry models. In other words, statistically significant sequence similarity indicates common ancestry.
In the end, I think Theobald has actually shown that different proteins (that are not significantly similar) are more likely to have different origins than to have a common origin. He has tested the hypothesis of universal common ancestry of all proteins. He also formalized Fitch's old test of protein homology from 1970. In a classic paper in Systematic Zoology, Fitch argued
One can maintain convergence in the cytochrome c gene as a logical possibility only by going all the way and assuming that there must have been a very large number of origins (perhaps as many as 24) to the 24 cytochromes c that were analyzed in this study. But if such a position is to be advocated, one must also explain how so many independently arising genes should have, by themselves, led to a phylogeny of these species (Figure 5) which is so similar to the phylogeny biologists have produced using other characters. The explanation will become more tedious as other genes produce similar results until, like the geocentric view of the solar system, it collapses under the burden of epicycles of epicycles.In other words, we may infer that protein similarity is the result of common ancestry because protein similarities form a single, sensible tree. Just like Theobald showed that protein evolution models that include all similar proteins on a single tree are preferred to those that put them on separate trees.
Is this a test of universal organismal common ancestry vs. independent creation? Well, there's the rub. Steve Matheson has of late lamented the ID strategy of comparing design to random origins, since this is a false dichotomy. I think a similar response may apply here. Theobald's test compares one tree to separate trees for proteins with significant sequence similarity, and he finds that the one tree is preferred. As I see it, all he's tested is whether the similarity of proteins can be best described by a single tree or by multiple trees. The alternative model of multiple trees is only favored when there is no sequence similarity. What has not been tested is whether there could be multiple origins of similar proteins that were created to be similar, and that's really the question, isn't it? Obviously humans and bacteria have proteins that share significant sequence similarity. No one's questioning that. A correct alternative model would have to assess the probability of created similarity vs. evolved similarity. Impossible? Maybe ...
Here's the way Darwin put this general argument in his Essay of 1844:
I must here premise that, according to the view ordinarily received, the myriads of organisms, which have during past and present times peopled this world, have been created by so many distinct acts of creation. It is impossible to reason concerning the will of the Creator, and therefore, according to this view, we can see no cause why or why not the individual organism should have been created on any fixed scheme. That all organisms of this world have been produced on a scheme is certain from their general affinities; and if this scheme can be shown to be the same with that which would result from allied organic beings descending from common stocks, it becomes highly improbable that they have been separately created by individual acts of the will of a Creator.I've discussed this passage elsewhere, but here I'd like to emphasize the conditional "if this scheme can be shown to be the same with that which would result from allied organic beings descending from common stocks." I think one possible test of this claim would be to determine whether proteins give a consistent phylogeny, or to test how much phylogenetic "signal" there is in proteins. If proteins do not show a scheme that is consistent with "allied organic beings descending from common stocks," that would be a highly relevant finding. It wouldn't falsify universal common ancestry (since there's always gene transfer, duplication/deletion, rate variation, etc. to explain inconsistent phylogenies), but it would certainly open the door to alternative explanations of similarity not based on common ancestry.
As I've said before, I suspect that most proteins do not give a consistent phylogenetic signal. At the very least, if the exception has become the rule, shouldn't we be re-examining the rule?
Feedback? Email me at toddcharleswood [at] gmail [dot] com.
Theobald. 2010. A formal test of the theory of universal common ancestry. Nature 465:219-222.
Fitch. 1970. Distinguishing homologous from analogous proteins. Systematic Zoology 19(2):99-113.
Photo: Nature